Background: Prediction of patient outcomes following allogeneic hematopoietic stem cell transplantation (HSCT) remains a tenacious problem. An important limitation of current prediction models is the heterogeneity in outcome even among similar cases. We introduce a novel approach to individualizing estimates of leukemia-free survival (LFS) in acute leukemia patients undergoing haploidentical (haplo) HSCT.
Methods: Data were obtained from the registry of the European Society for Blood and Marrow Transplantation for all cases of haplo HSCT for acute leukemia performed between 2011 and 2017. Patients receiving ex-vivo T-cell depleted grafts were excluded. Acute myeloid leukemia patients were classified by clinical disease ontogeny (de novo vs. secondary), cytogenetics, and FLT3-ITD/NPM1mut status; acute lymphoblastic leukemia patients by disease status and the presence of the Philadelphia chromosome. Common patient and transplantation parameters including recipient age, Karnofsky performance status (KPS), time from diagnosis to transplantation, conditioning and graft-versus-host disease (GvHD) prophylaxis were included. Data were split into training and geographic validation sets.
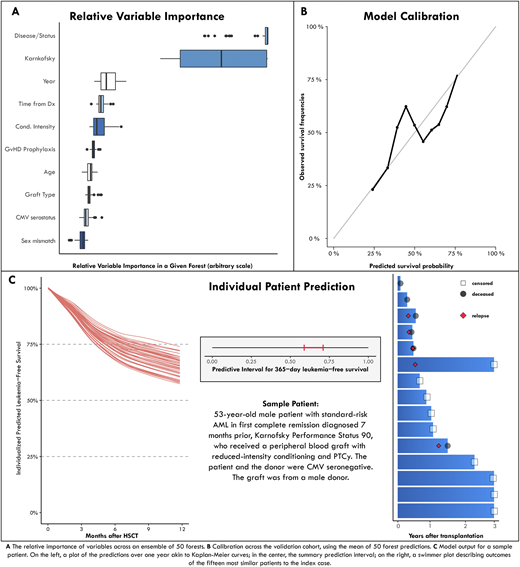
Results: A total of 2,001 patients was included in the training set and another 270 in the validation cohort. In the training set, the median age was 50 years; 68% of patients were in complete remission, and 69% had a KPS ≥ 90; 87% received post-transplant cyclophosphamide and 13% antithymocyte globulin for GvHD prophylaxis. To provide the clinician insight on outcomes of similar patients, we developed a descriptive tool to visually explore outcomes of cases with comparable features. We next generated 50 random survival forest models for the prediction of 1-year LFS. In contrast to single point-estimates, the ensemble of 50 models generates a prediction interval accounting for predictive uncertainty. There was heterogeneity of variable importance between models, with either disease status or KPS leading in all models (Figure A). The model was well calibrated (Figure B); the median c-statistic was 0.64 on the validation set. An online interface presents the individual outcomes of the fifteen patients most similar to the index case, the prediction interval, and a visualization of all 50 survival forest predictions. Predictions for a sample patient are shown in Figure (C).
Conclusions: We present the first system for individualized prediction of leukemia-free survival following T-cell replete haplo transplantation. A key, novel component of the model, distinguishing it from standard risk scores, is that it provides a measure of predictive certainty. This is essential for judging the robustness of prediction. Our approach is applicable to other clinical settings and can be used for designing risk-guided interventions and for informing patients and clinicians.
Labopin:Jazz Pharmaceuticals: Honoraria. Blaise:Jazz Pharmaceuticals: Honoraria. Sica:F. Hoffmann-La Roche Ltd: Other: All authors received support for third-party writing assistance, furnished by Scott Battle, PhD, provided by F. Hoffmann-La Roche, Basel, Switzerland., Research Funding. Mohty:Stemline: Consultancy, Honoraria, Research Funding, Speakers Bureau; Takeda: Consultancy, Honoraria, Research Funding, Speakers Bureau; Jazz Pharmaceuticals: Consultancy, Honoraria, Research Funding, Speakers Bureau; Janssen: Consultancy, Honoraria, Research Funding, Speakers Bureau; Sanofi: Consultancy, Honoraria, Research Funding, Speakers Bureau; Novartis: Consultancy, Honoraria, Research Funding, Speakers Bureau; BMS: Consultancy, Honoraria, Research Funding, Speakers Bureau; Celgene: Consultancy, Honoraria, Research Funding, Speakers Bureau; Amgen: Consultancy, Honoraria, Research Funding, Speakers Bureau; GSK: Consultancy, Honoraria, Research Funding, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal